Recently, a study led by the University of Toronto in Canada and uk-silicon intelligence showed for the first time the potential of quantum computing and artificial intelligence to transform the drug discovery process. In this study, scientists combine quantum computational models with classical computational models and generative artificial intelligence to explore a wider range of chemical possibilities through the training, generation, and screening of large data sets, and to explore the potential applications of quantum computational models, discovery of novel molecules targeting the ‘non-druggable’ cancer kinesin KRAS. The research, published in Nature Biotechnology on January 22, has also been supported by St. Jude Children’s Research Hospital and other institutions.
KRAS mutations are one of the most common mutations in cancer, occurring in about a quarter of human tumors. KRAS mutations cause cells to proliferate uncontrollably, which can lead to cancer. Despite the vast prevalence and impact of the KRAS mutation, only two drugs that specifically target the mutant KRAS have been approved by the US Food and Drug Administration (FDA) . Moreover, clinical data show that these drugs extend patients’ lives by only a few months compared with conventional chemotherapy. There is an unmet need to improve KRAS-targeted therapy to bring more benefits to cancer patients.
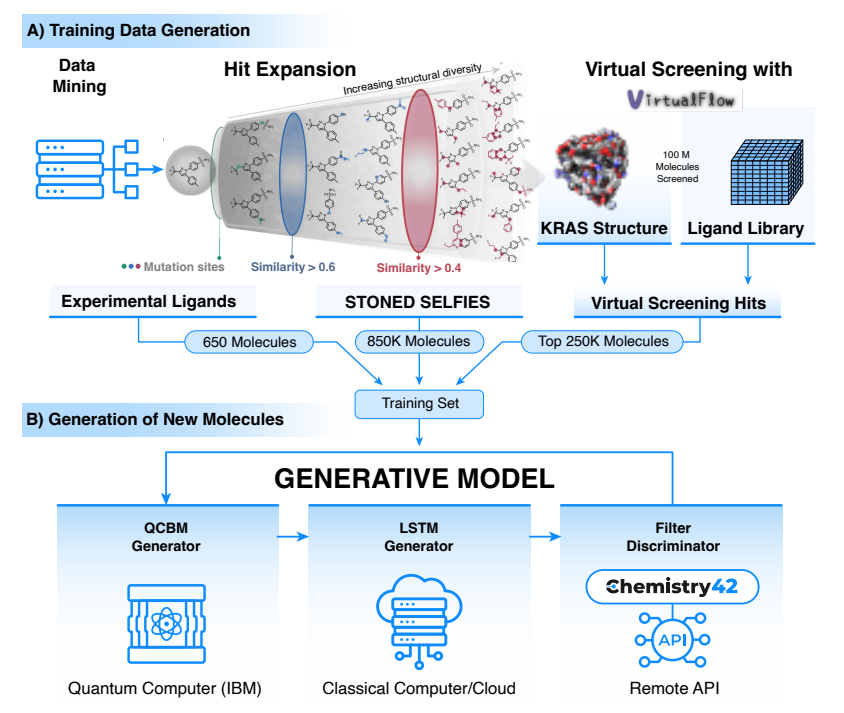
Hybrid model training and molecular generation process
In this study, in order to generate potential novel KRAS inhibitors, the researchers proposed a quantum-classical hybrid framework model combining the quantum variational generation model (QCBM) and the long short-term memory network (LSTM) , which can be used to generate novel KRAS inhibitors, combining quantum computation with classical computation to design new molecules. They first trained the hybrid model with a custom dataset of 1.1 million molecules, including 650 previously experimentally validated KRAS-blocking molecules, obtained from the literature, and compared the performance of the hybrid model with that of the classical KRAS-blocking model, using the STONED-SELFIES algorithm to generate 850,000 analogs with reference to existing KRAS inhibitors, and 250,000 molecular data obtained through the open-source very large virtual screening platform VirtualFlow, we identified 100,000 KRAS inhibitors, 100,000 KRAS inhibitors, 100,000 KRAS inhibitors, and 100,000 KRAS inhibitors, these trainings help to optimize the integrated model.
Next, the team generated a million candidate molecules using the hybrid model. In this process, QCBM, as a quantum generation model, uses quantum circuits to learn complex probability distributions, thereby generating new samples similar to the training data. In addition, it also acts as a prior throughout the process, guiding LSTM to generate new ligand samples. As a classical model, LSTM processes the sequence data of chemical structure and generates new molecular sequences based on it. This combination enables the generated molecules to obtain complex probability distributions from quantum models and to be serialized through classical models, thereby improving the quality and diversity of the generated molecules.
The research team further filtered the molecules using Chemistry42, the generative AI engine of uk-silicon Intelligence, which includes multi-dimensional evaluation metrics such as drug-like screening, molecular docking ranking, and synthetic accessibility, and 15 of the most promising candidates for laboratory testing were identified. After the wet test, two of the 15 candidate molecules stood out with novel structures and no significant non-specific cytotoxicity.
Among them, ISM061-018-2 showed better binding ability to target proteins than other molecules. In addition to wild-type KRAS, ISM061-018-2 showed dose-dependent inhibitory activity against five other common mutant KRAS as well as wild-type HRAS and NRAS, respectively, this result demonstrates its potential as a pan-RAS inhibitor with an entirely new structure. Another compound with pan-Ras inhibitor potential, ISM061-022, was also shown to exhibit enhanced inhibitory activity against specific mutant KRAS (G12R and Q61h) relative to ISM061-018-2.
“Traditional drug discovery methods rely on screening existing compounds libraries to find compounds that are active against specific target proteins,” said study co-author Dr Igor Stagljar, a professor of biochemist and molecular genetics at the University of Toronto. These methods are costly, time-consuming, and involve complex processes for compound management. With innovative methods, much of the screening can be done in the cloud, without the need for physical space to store chemical libraries or for robots to do large-scale screening.”
Notably, while these results demonstrate the potential of quantum computing to accelerate the early stages of drug discovery, they do not demonstrate that molecules discovered with this approach are more effective than those discovered with classical methods. Dr Al án Aspuru-Guzik, professor of chemistry and Computer Science at the University of Toronto, Canada, who led the study, said: “It is exciting to work on the interdisciplinary convergence of medicinal chemistry, quantum computing and artificial intelligence. This pioneering study demonstrates that quantum computers can be incorporated into modern artificial intelligence-driven drug discovery processes and successfully find molecules that interact with biological targets. This is a proof-of-principle study, but it does not provide any indication that quantum advantage is significant. As quantum computers become more powerful, our algorithms are expected to get better and better.”
Building on their success with KRAS, the researchers are now applying their quantum-classical hybrid model to other“Undruggable” proteins, such as KRAS, which are often small, the lack of a surface profile makes it difficult for molecules to bond with it. The team also plans to use the hybrid model to further optimize the design of two promising KRAS inhibitors in animal models.
Dr Alex Zhavoronkov, co-author of the paper and founder and chief executive of silicon intelligence, said: “Up to 85 per cent of human proteins are considered ‘undruggable’ , with smooth surfaces and lack of convenient pockets for small molecule drug binding. This is one of the big challenges in developing new treatments for cancer, where AI has a unique advantage. The collaboration between the University of Toronto and uk-silicon intelligence is a good example of how start-ups and research institutes can use their expertise to advance technology and better work together for the long-term health of humanity.”