How to accurately identify specific bacteria in a sea of bacteria and induce them to release specific molecules to activate the immune response? On June 13, reporters learned from the Chinese Academy of Sciences Shanghai Institute of Nutrition and Health that the research group of the institute’s researcher Qian youcun and the research group of Song Xinyang, researcher of the Center for Excellence and Innovation in Chinese Academy of Sciences Molecular Cell Science, the molecular mechanism of specific binding of APOL9 protein to bacteroidetes in the gut has been found. The APOL9 protein acts as a trained“Bacterial diplomat” to help maintain the immune homeostasis of the gut by building delicate cooperation with specific bacteria. The findings were published in the international journal Nature.
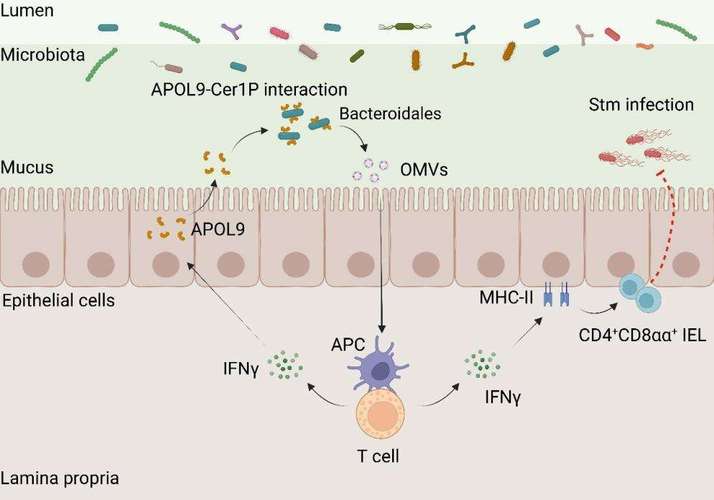
As the first barrier for host-microbe contact, intestinal epithelial cells not only constitute a physical barrier, but also regulate microbiota structure and function by secreting multiple functional proteins. However, how these secreted proteins accurately recognize specific bacteria and the molecular mechanisms that shape the host immune environment through commensal bacteria have been difficult to study. Unlike traditional antimicrobial proteins, APOL9 does not kill the target bacteria, but induces them to release outer membrane vesicles (OMVs) , Qian said. These nano-sized vesicles are loaded with bacterial molecules that can be captured by the host’s immune system and used to boost immune defenses. Specifically, OMVs can activate the interferon-? signaling pathway and increase the expression of MHC-II molecules on the surface of intestinal cells, thereby training a special class of CD4 + CD8 ?? + T cells to maintain intestinal immune homeostasis.
To test the physiological function of APOL9, the team constructed Apol9 knockout mice and infected them with salmonella, an intestinal pathogen. During the experiment, the mice showed uncontrolled spread of bacteria and a significant increase in mortality. When these mice were supplemented with OMVs, the symptoms of infection were significantly improved and the immune response was significantly enhanced.
This groundbreaking study is the first to demonstrate that host proteins can trigger beneficial immune responses by recognizing bacterial lipid markers. The research also reveals a new paradigm in which the host actively shapes the microbiota — not by passively tolerating microbes, but by actively “Talking” to molecules to Dynamic equilibrium the gut’s microbial ecology. This discovery opens up new avenues for the development of next-generation therapies for the co-regulation of“Microbiota-immunity”.